 Run Analysis
Documentation
Contact us
Run Analysis
Documentation
Contact us

OpenVar is the first tool for genomic variant annotation and functional effect prediction supporting deep open reading frame (ORF) annotation and polycistronic annotation of Human, Mouse, Rat and Fruit fly transcripts.
OpenVar builds on the well-known and extensively used SNPEff tool (Cingolani et al., 2012), but also offers the possibility to predict variant effect in alternative ORFs as defined in OpenProt (Brunet et al., 2019).
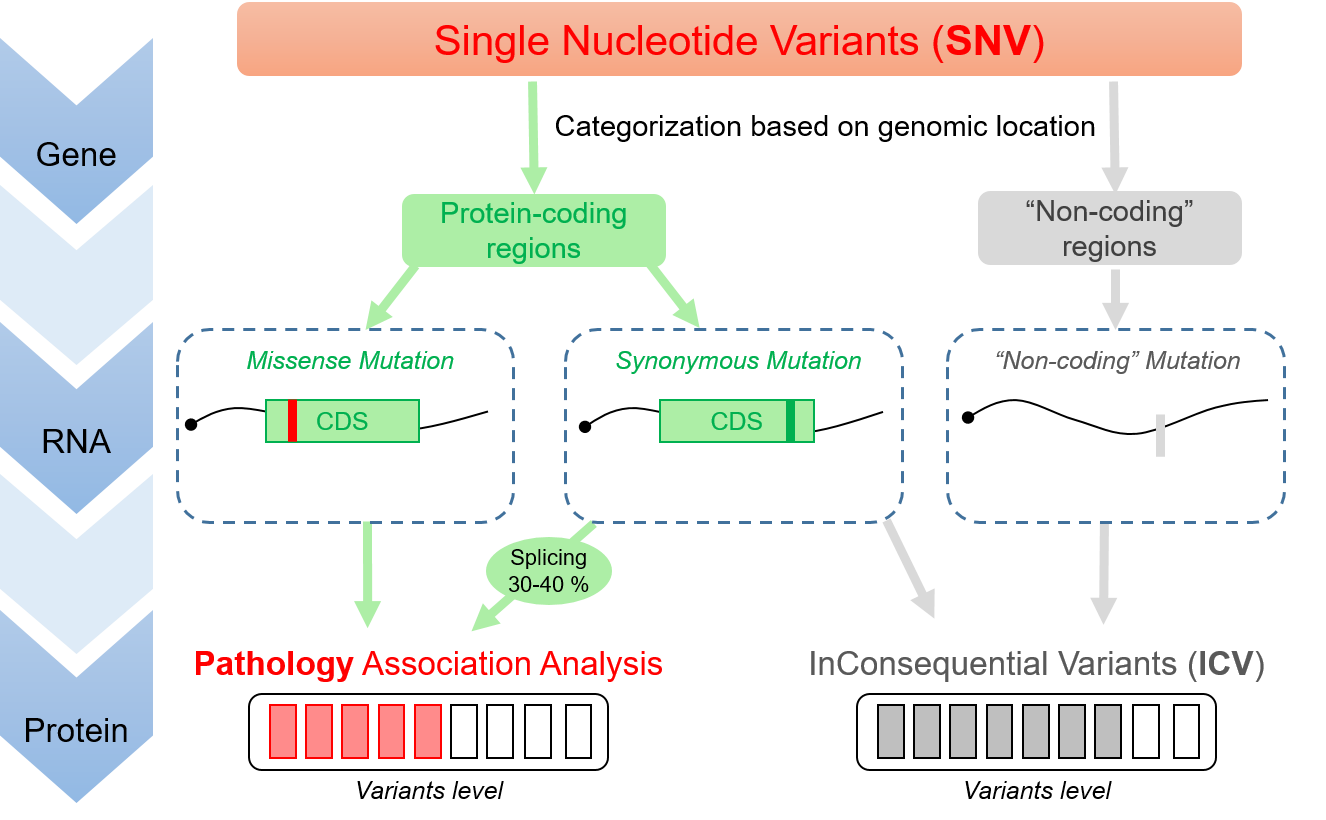
With current genome annotations, about 75% of genomic variants are deemed inconsequential (ICV). These ICV correspond to variants falling in allegedly non-coding regions, such as UTRs or ncRNAs, or yielding synonymous mutations (silent). Yet, some ICV have been strongly associated with pathologies and only about 30-40 % can be explained with hypotheses formulated under a monocistronic annotation of the human genome.
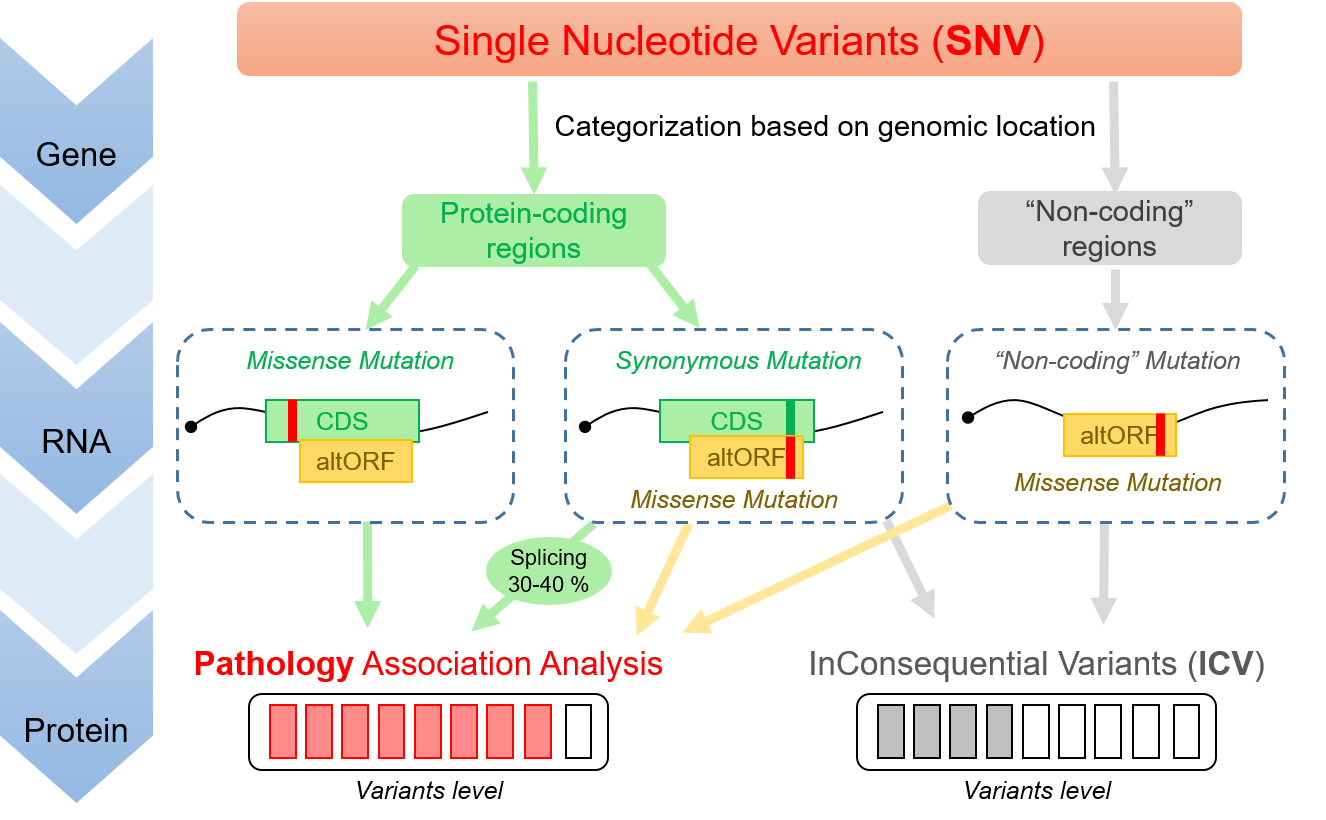
By allowing annotation of multiple ORF per transcript (polycistronic annotation), OpenProt provides a more complex yet more comprehensive view of the coding potential of human genes. Early studies suggest that up to 30% of ICV could be explained by their consequence on a currently unannotated protein.
38% of Mendelian phenotypes still have no known molecular basis (OMIM).
Up to 30% of synonymous mutations associated to cancers alter unannotated ORF (Brunet et al, 2018).
80% of rare diseases are thought to be of genetic origin, yet only 39% have a described gene association (Ferreira, 2019).